Introduction
The isolation of plasmid DNA from bacteria is an essential molecular biology technique, used to produce template DNA for desired downstream reactions. Plasmid isolation methods are simple, however, high-throughput plasmid DNA extraction has proved problematic, with issues including low yield and genomic DNA (gDNA) contamination. An automated, high-throughput, multiwell-based plasmid extraction method, with a yield appropriate for downstream applications and free from gDNA, will greatly benefit high-throughput laboratories, where plasmid extractions is often a bottle-neck.
96-well, silica filter-plate plasmid extractions kits are available commercially. In these methods, bacteria cells are harvested and lysed by alkaline lysis, the cellular debris is removed, and the extracted DNA is subsequently collected by binding to a silica membrane. Membranes are then washed, prior to the elution of purified DNA. Here, we describe a method developed at SynbiCITE’s London DNA Foundry which uses the CyBio FeliX pipetting platform to automate a 96-well, silica filter-plate based plasmid DNA extraction protocol.
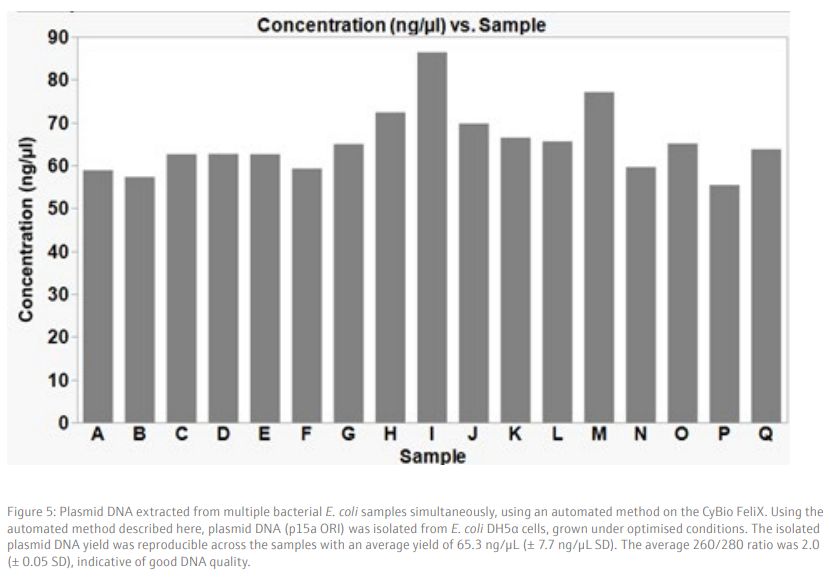
The use of the CyBio FeliX pipetting platform allows 96 samples to be processed simultaneously, in approximately 1,5 hours, with the flexibility to process multiple plates at one time. The compact platform reduces the bench space needed for an automated plasmid isolation system in a laboratory and, in combination with a bench top centrifuge and robotic arm, can enable a fully automated system for plasmid extraction. Using this method, an average yield of 65 ng/µL plasmid DNA is isolated from bacterial cultures, with low variability between the samples (+/- 7.7 ng/µL SD) and in an elution volume of 50 µL. The plasmid DNA samples are also high quality, suitable for downstream applications such as sequencing or transformation.

Materials and Methods
Reagents and Instrumentation
 Sample preparation
Sample preparation
- Grow bacterial clones, containing plasmid DNA of interest
- Inoculate single clones into a 96-well, 2 mL, square-well, plate containing 1.2 mL/well Terrific broth (TB) with appropriate antibiotic(s)
- Grow cells overnight shaking, 37° C
- N.b. use high copy number plasmid for increased yield
Method
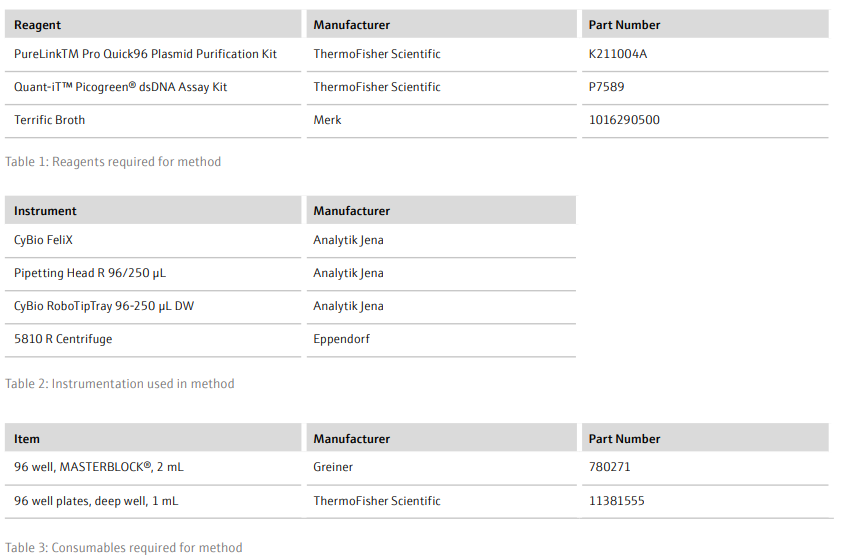
- Prepare all buffers as described in the manufacturer’s instructions and dispense into 96-well, 1 mL plates according to the volumes in Table 4 (volumes given are appropriate for 1 x 96-well plasmid extraction)
- Centrifuge the plate containing overnight bacterial cultures at 2250 x g, 10 minutes
- Remove the supernatant, ensuring cell pellets remain intact
- Add RoboTipTrays (x5), buffer plates (x5), sample plate (containing cell pellets) and the ‘Clarification’ plate (on top of a 96-well, 1 mL plate) to the deck of the CyBio FeliX. For example, as detailed in Figure 2
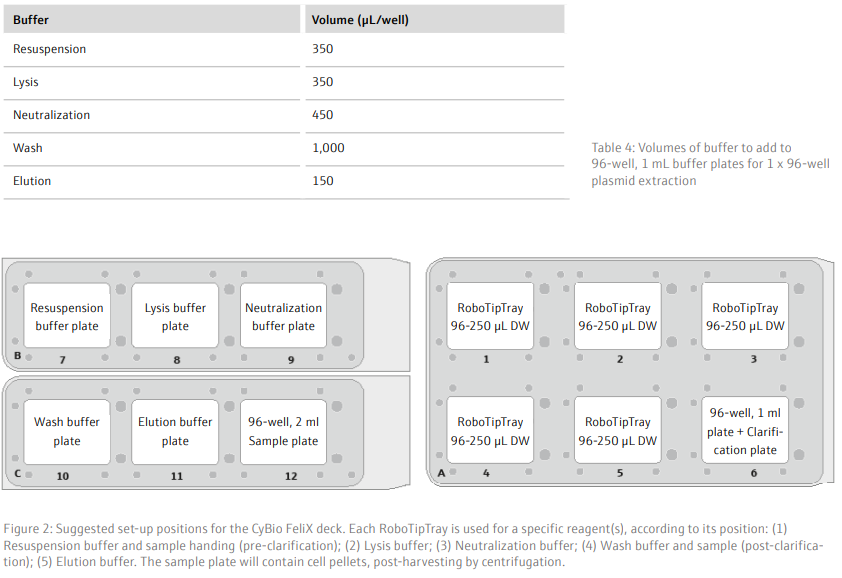
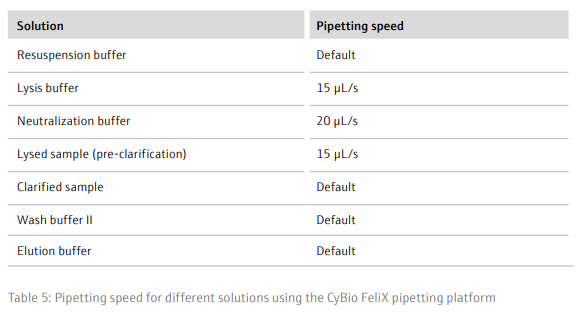
- Using the CyBio FeliX, add 250 µL/well resuspension buffer to the sample plate and mix thoroughly (with the same set of tips) for 3 minutes to resuspend the cell pellets
- Add 250 µL/well lysis buffer to the sample plate
- Mix using the sample tips for 4 minutes to lyse the cells
- Add 350 µL/well neutralization buffer to the sample plate, to stop acidification of the DNA
- Mix using the sample tips for 3 minutes
- Transfer the total volume of lysed sample (850 µL/well) to the Clarification plate (provided in kit), using the sample tips
- Centrifuge the Clarification plate, on top of 1 mL, deep-well collection plate at 2250 x g, 2 minutes. The sample will pass through the clarification filter, transferring the DNA to the collection plate below and removing the cell debris
- Discard the Clarification plate and return the 1 mL, deep-well collection plate to the top deck of the FeliX (position 12, replacing the 96-well, 2 mL Sample plate)
- Add the ‘Filter’ plate (provided in kit), on top of a clean 1 mL, deep-well collection plate to the bottom deck of the FeliX (position 6)
- Transfer the entire volume of the clarified sample (850 µL/well) to the Filter plate, using clean sample tips
- Centrifuge the Filter plate, on top of a 1 mL, deep-well collection plate at 2250 x g, 2 minutes. The DNA will now be bound to the silica membrane in the Filter plate
- Discard the flow-through solution from the 1 mL, deep-well collection plate
- Place the Filter plate back on top of the 1 mL, deep-well collection plate on the bottom deck of the FeliX (position 6)
- Add 900 µL/well wash buffer to the Filter plate, using the sample tips (n.b. wash buffer plate must be prepared fresh each time)
- Centrifuge the Filter plate, on top of the 1 mL, deep-well collection plate at 2250 x g, 2 minutes. The DNA will remain bound to the silica membrane in the Filter plate
- Discard the flow-through solution from the 1 mL, deep-well collection plate
- Centrifuge the Filter plate, on top of the empty 1 mL, deep-well collection plate at 2250 x g, 10 minutes, to get rid of any residual wash buffer. The DNA will remain bound to the silica membrane in the Filter plate
- Place the Filter plate on top of the ‘Elution’ plate (provided in kit) on the bottom deck of the FeliX, (position 6)
- Add 50 µL/well elution buffer to the Filter plate
- Incubate the plate at room temperature for 4 minutes
- Centrifuge the Filter plate, on top of the Elution plate at 2250 x g, 2 minutes, to elute the DNA from the silica membrane
- Seal the Elution plate containing the purified plasmid DNA samples with a foil seal and store at 4° C (short term) or -20° C (long term)
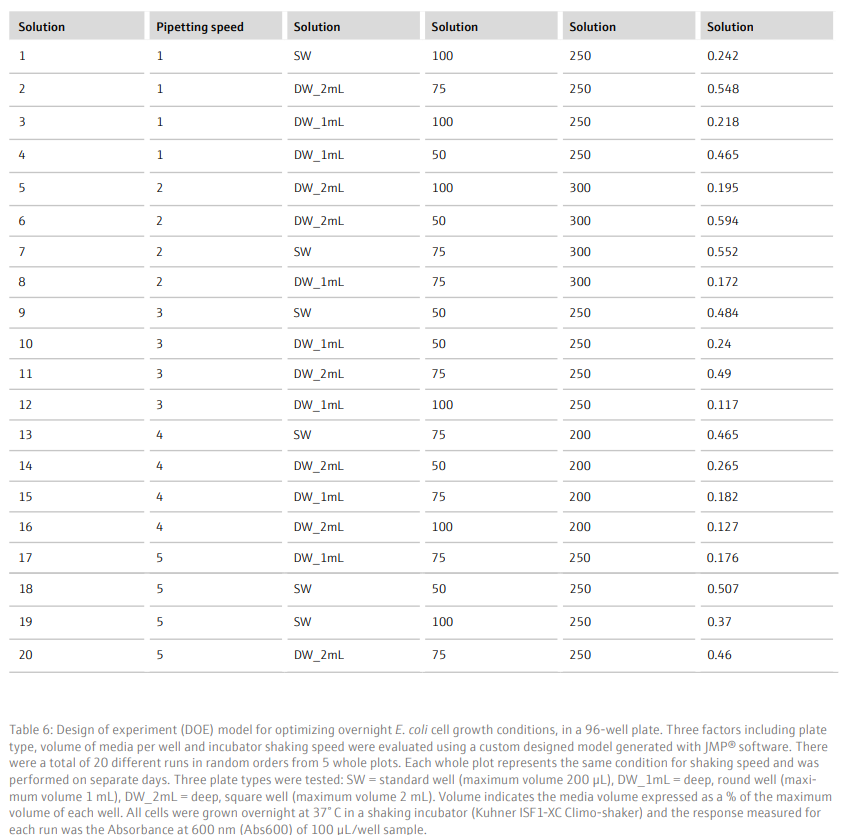
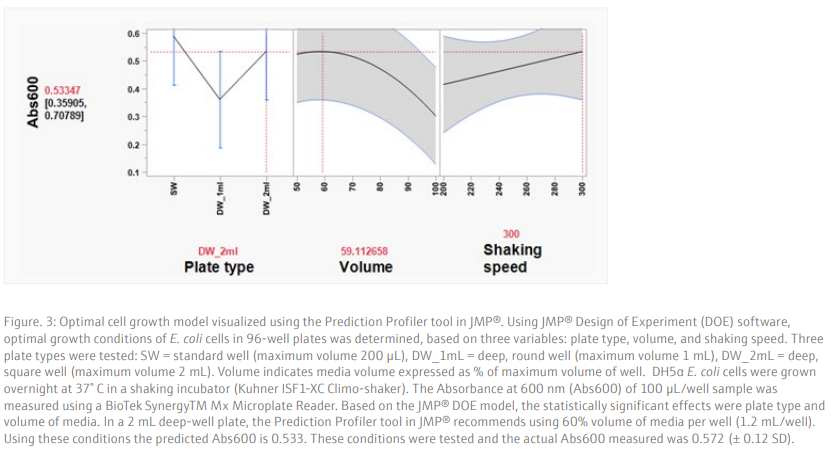
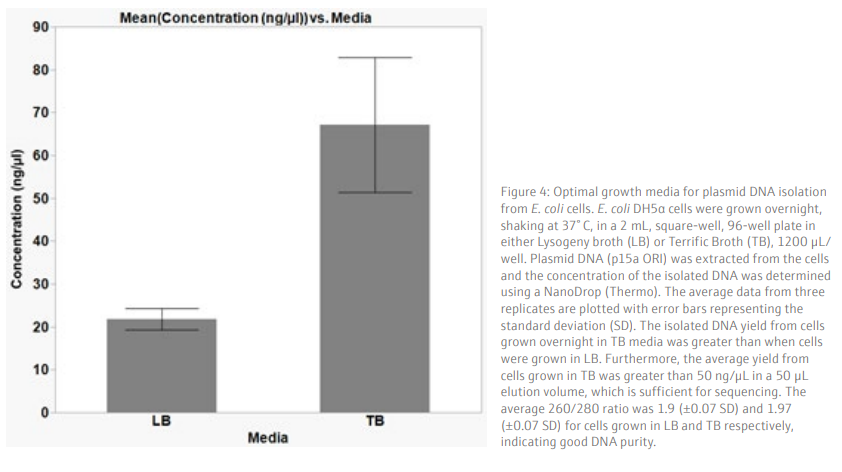
Results and DiscussionTo achieve a good yield from plasmid DNA isolation, it is important to grow bacteria cells optimally prior to running the method. We used a Design of Experiments (DOE) approach to optimize the overnight growth of DH5α Escherichia coli (E. coli) cells at 37° C. The amount of cell growth was determined by measuring the Absorbance of 100 µL/well culture at 600 nm (Abs600). Using a custom designed model, generated in the JMP® software, we found that the optimal conditions for cell growth was to use a 2 mL, square-well, 96-well plate, containing 60% volume of media per well (1.2 mL/well) (Table 6 and Figure 3). To note, high Abs600 values were also obtained when cells were grown in a standard depth, 96-well plate. However, due to the reduced volume capacity of the wells in these plates, the total number of cells was not sufficient for plasmid DNA extraction. We found that shaking speed did not have a significant effect on cell growth. Furthermore, we determined that the growth of cells in Terrific Broth (TB) improved the yield of isolated plasmid DNA, as compared to Lysogeny broth (LB) (Figure 4). The growth of bacteria cells under optimized conditions is essential for obtaining a good yield of plasmid DNA. The yield obtained under the optimized conditions was greater than 50 ng/µL (Figure 4), in a 50 µL elution volume, which is sufficient for downstream applications such as sequencing [1].
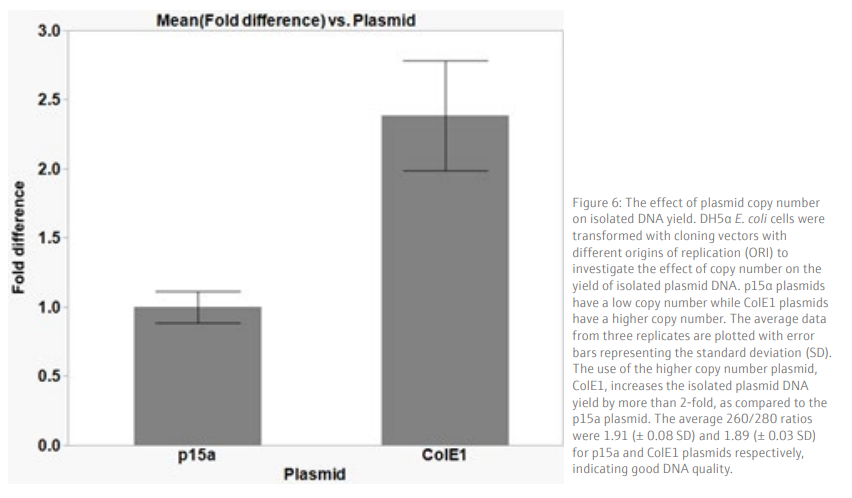
We demonstrate that the plasmid isolation method described here gives a reproducible yield from samples processed simultaneously in a 96-well plate (Figure 5). High quality DNA, with an average yield of 65.3 ng/µL (+/- 7.7 SD), in a 50 µL volume is obtained from a low copy number plasmid (p15a ORI), which is sufficient for downstream applications such as sequencing or transformation into bacteria cells. As expected, the use of a higher copy number plasmid generates a higher yield of isolated plasmid DNA (Figure 6). Therefore, the use of high copy number plasmids offers an option for higher yield of plasmid DNA, if required.
The method described here can be used to process a multiwell plate of 96 samples simultaneously, using the CyBio FeliX pipetting system. If used in combination with a robotic arm and an integrated centrifuge with sufficient depth for the filter plates, this method can be fully automated and adapted to process multiple 96-well sample plates at one time.
Conclusion
As the demand for higher throughput in molecular biology laboratories increases there is a necessity for the implementation of essential techniques, such as plasmid DNA isolation, on a high-throughput, automated scale. Here, we aimed to develop a method for the high-throughput isolation of plasmid DNA from bacterial cell cultures, which can be fully automated. We demonstrate that this method isolates plenty of high-quality, plasmid DNA from bacterial cultures, at yields suitable for downstream applications. The DNA yield is highly dependent on the quality of the overnight growth of bacterial cultures and therefore we outline optimised growth conditions which are essential for the success of this method. The use of the compact CyBio FeliX pipetting platform for high-throughput plasmid DNA isolation offers an option to enable molecular biology laboratories to keep up with demand and to overcome a common bottleneck in laboratory workflow.